On May 8th, the international academic top journal “Nature Genetics” online published a long article about the research of the germplasm resources of north China crops, which utilized the national key laboratory of the Ministry of Education of P.R.C., the teachers’ team led by Prof. Ma Zhiying, united with Institute of Cotton, Chinese Academy of Agricultural Sciences and other units, completed the latest research outcome “Resequencing a Core Collection of Upland Cotton Identifies Genomic Variation and Loci Which Influencing Fiber Quality and Yield”. The study, for the first time, resequenced the genome of 419 upland cotton core germplasms from China, the United States and Australia, (representing 7362 samples of upland cotton from China's cotton germplasm bank). The study is a breakthrough in the field of cotton genomic variation, genetic fiber traits, important character phenotype of cotton core germplasm, genome variation & molecular marker identification and new gene discovery, which provides an important basis for deepening the research on crop genome, and more accurate marks and gene resources for targeted breeding of cotton. Ma Zhiying, Wang Xingfen, Zhang Yan, Zhang Guiyin, Wu Liqiang, Li Zhikun, from HEBAU; He Shoupu, Sun Junling, from China Cotton Institute; Liu Zhihao, from Novogene Corporation are the first authors that contribute equivalent to the paper. Ma Zhiying, Wang Xingfen, from HEBAU; Du Xiongming, from China Cotton Institute; Tian Shilin, from Novogene Corporation are the corresponding authors of the paper.
Upland Cotton takes more than 90% of the world's cotton planting because of its wide adaptability and high yield. Currently, 95% of the cotton produced in China comes from upland cotton. Long-term natural selection and artificial breeding have produced a large number of upland cotton germplasm resources, and it is an important research work to dig deep into the genome variation of core germplasm. And with the people's increasing demand and the improvement of spinning and weaving process, it requires higher on the quality of cotton fiber, and deepens the research of molecular basis of phenotypic variation of germplasm resources and excellent loci of genetic variation, which can realize the effective selection and improvement of the important traits such as: cotton quality and yield. Prof. Ma Zhiying led the team of cotton over six years, through actual environment plant identification and carried out the comprehensive analysis and summary at the DNA level based on existing breeding works. The results of research provides the theory basis and molecular breeding gene resources for cotton breeding technology.
The study shows that the average depth of genomic resequence of 419 samples of upland cotton core germplasm were 6.55 times, identified 3665030 single-base polymorphism (SNP), it is found that genetic diversity was higher than the local places of upland cotton varieties and the modern improved varieties. It is also reported that the core germplasm group provided a relatively broad genetic basis for breeding. By comparing modern varieties with early varieties, it is found that the diversity of nucleotide was reduced by 8.6%, which provided the direct molecular evidence for the reduction of genetic diversity of cotton by artificial domestication. By comparing the core germplasm and upland cotton wild genome-wide SNP variation of functional genes, it is the first time to be found 23876 genes have no SNP variation, which indicates these genes in the process of long-term domestication are highly conservative. SNP increases and decreases are respectively found in 33899 and 6957 genes ,which suggested that these genes should be given priority attention in breeding improvement.
The study was conducted in 12 environments of 6 sites in the Yellow River basin, the Yangtze river basin and the northwest inland three cotton areas in 2014-2015. The quality and yield characters of 13 fibers, such as fiber length, strength, weight and clothing were identified, and nearly 200,000 phenotype data were obtained. Based on the total genomic association analysis of 3,665,030 SNPS, a total of 11,026 SNPS that were significantly associated with 13 traits were identified.Among them, A large number of loci of SNP can be repeatedly tested in at least 3 environments, and for the first time, the detection of SNP associated with fiber quality is far more than that of fiber yield SNP. Compared with the chromosome of a subgroup, the chromosome of D subgroup is more related to SNP. It identifies the key chromosomes distributing in different genetic loci.
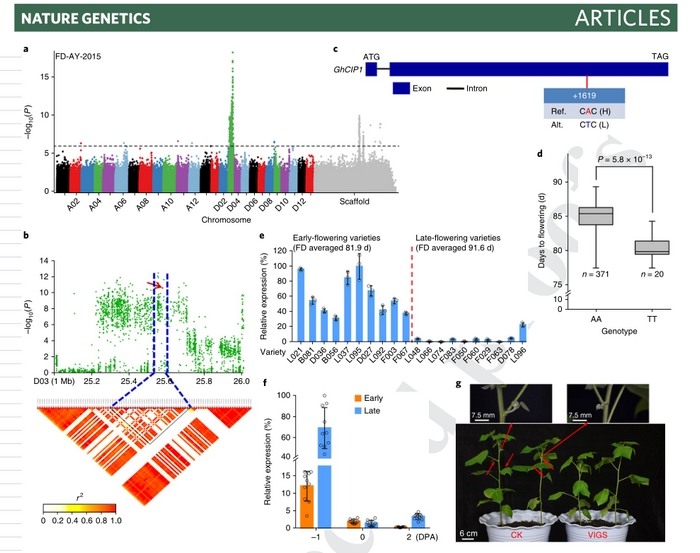
The study identified 7,398 genes. Combined with four different varieties of cotton fiber development in different periods of the transcriptome analysis, it was found that 3,089 genes were expressed at different stages of fiber development, and there were many new genes related to flowering, fiber length and fiber strength. By gene quantitative expression through different haplotype varieties, gene over expression and gene silencing test, for the first time, the study identified a new gene “GhCIP1, GhUCE”, fiber length new gene “GhFL1, GhFL2” and 1 fiber strength new gene “Gh_A07G1769”.
The study also identified 5,753 excellent loci from 7,383 significant single nucleotide polymorphisms. The frequency of excellent loci of wild species, early varieties and modern varieties was further evaluated, and the frequency of all excellent loci was significantly increased during domestication.This finding is consistent with the improvement of fiber yield in the process of domestication as the selection target, and in modern breeding takes the quality of fiber as well as yield as the improving target, which reflects an important trait phenotypic selection to preserve the molecular evolution of an excellent genetic loci.
The research is supported by the National Modern Agricultural Industry Technology System, Science and Technology Support Key Plan in Hebei Province, National Key Science and Technology Project, and National Key Research and Development Plan.
It is reported that the cotton team led by Prof. Ma Zhiying of HEBAU has successively published scientific achievements in international top academic journal “Nature Genetics” and “Nature Biotechnology”as the first author at the same contribution to the first author respectively in 2014 and 2015. The team after more than 30 years of development has formed an cotton research team which caught international influence, and 1 item won the HeLiangHeLi Scientific and Technological Innovation Fund, 1 item of National Scientific and Technological Progress in second prize, 4 Scientific and Technological Achievement of Hebei Province in first prize, 1 National Teaching Masters’ Award, 1 National Teaching Achievement Award in second prize.
Reporter: Yao Yunxiao Editor: Shi Chunxiang
Translated by Ren Xiaokun and Proofread by Ran Longxian